Difference between revisions of "Slicer3:JavaBasedChainCLMsEngine"
From NAMIC Wiki
(New page: = Java Based Chain CLMs Engine = == Introduction == * Java application which behaves as a regular Slicer3 module. * Requires as its first parameter the location of a chain descriptor. * ...) |
|||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | = | + | = Chain CLMs Engine = |
== Introduction == | == Introduction == | ||
| − | * Java application which behaves as a regular Slicer3 module. | + | * Java application which behaves as a regular Slicer3 module (JVM 1.5 or higher required). |
* Requires as its first parameter the location of a chain descriptor. | * Requires as its first parameter the location of a chain descriptor. | ||
* The rest of its parameters are the usual Slicer3 modules parameters. | * The rest of its parameters are the usual Slicer3 modules parameters. | ||
| Line 18: | Line 18: | ||
* For each chain module desired create: | * For each chain module desired create: | ||
| − | ** A "X-Chain.xml" chain descriptor file(see "Chain Descriptor" section for details). | + | ** A "X-Chain.xml" chain descriptor file to be stored under the Slicer3 "Plugins" directory (see "Chain Descriptor" section for details). |
** A chain script module for "X-Chain.xml"; which should look like: | ** A chain script module for "X-Chain.xml"; which should look like: | ||
| Line 108: | Line 108: | ||
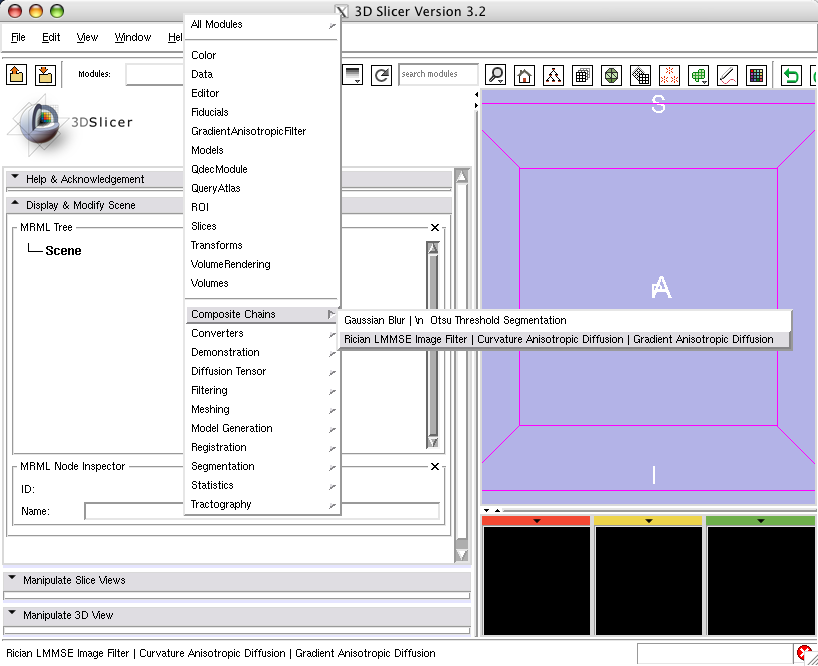
[[Image:Chain2GeneratedUI.png]] | [[Image:Chain2GeneratedUI.png]] | ||
| + | |||
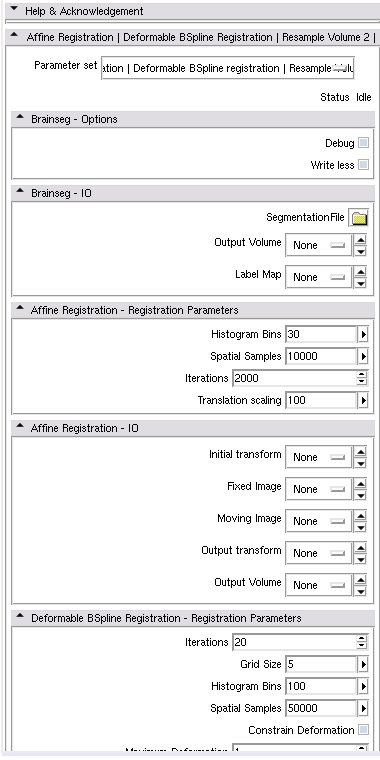
| + | * Chaining modules brainsegCLP, AffineRegistration, BSplineDeformableRegistration, ResampleVolume2, CortThickCLP | ||
| + | |||
| + | <?xml version="1.0" encoding="UTF-8"?> | ||
| + | <modules xmlns="http://www.na-mic.org/clm" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.na-mic.org/schemas/chainCLM.xsd"> | ||
| + | <module id="brainsegCLP"/> | ||
| + | <module id="AffineRegistration"/> | ||
| + | <module id="BSplineDeformableRegistration"> | ||
| + | <argument name="inputVolume" value-ref="AffineRegistration.ResampledImageFileName" /> | ||
| + | <argument name="InitialTransform" value-ref="AffineRegistration.OutputTransform" /> | ||
| + | <argument name="FixedImageFileName" value-ref="AffineRegistration.FixedImageFileName" /> | ||
| + | <argument name="MovingImageFileName" value-ref="AffineRegistration.MovingImageFileName" /> | ||
| + | </module> | ||
| + | <module id="ResampleVolume2"> | ||
| + | <argument name="InitialTransform" value-ref="BSplineRegistration.OutputTransform" /> | ||
| + | </module> | ||
| + | <module id="CortThickCLP"> | ||
| + | <argument name="SegmentationInput" value-ref="brainseg.SegmentationFile" /> | ||
| + | <argument name="ParcellationOption" value-ref="ResampleVolume2.outputVolume" /> | ||
| + | </module> | ||
| + | </modules> | ||
| + | |||
| + | [[Image:RegionalCorticalThicknessChain.jpg]] | ||
= Key Investigators = | = Key Investigators = | ||
| − | * UCSD | + | * UCSD: Marco Ruiz |
* Isomics: Steve Pieper | * Isomics: Steve Pieper | ||
* GE: Jim Miller | * GE: Jim Miller | ||
Latest revision as of 15:10, 5 January 2009
Home < Slicer3:JavaBasedChainCLMsEngineContents
Chain CLMs Engine
Introduction
- Java application which behaves as a regular Slicer3 module (JVM 1.5 or higher required).
- Requires as its first parameter the location of a chain descriptor.
- The rest of its parameters are the usual Slicer3 modules parameters.
- This java application is currently wrapped in a shell script; which would be the actual Slicer3 chain module.
- As such this script has to be deployed in the Slicer's Plugins folder.
- This script does pre and post processing before invoking the engine and hardcodes a chain descriptor.
Installation
- Unbundle the chain module engine right under your Slicer3 installation.
- Set the environmental variable SLICER_HOME to point to your Slicer3 installation. For example:
export SLICER_HOME=~/Slicer3.2-2008-06-02-darwin-x86
- For each chain module desired create:
- A "X-Chain.xml" chain descriptor file to be stored under the Slicer3 "Plugins" directory (see "Chain Descriptor" section for details).
- A chain script module for "X-Chain.xml"; which should look like:
#!/bin/sh
# Chain Slicer Module
export GWE_HOME=`ls -d -t $SLICER_HOME/gwe-*/ | head -1`
export GWE_FAT_JAR=`ls $GWE_HOME/lib/gwe-*-fatjar.jar`
rm -fdr $SLICER_HOME/chains
mkdir $SLICER_HOME/chains
java -Xmx512m -cp $GWE_HOME/conf:$GWE_FAT_JAR org.gwe.integration.slicer.chains.ChainCLMProxyApp $SLICER_HOME/lib/Slicer3/Plugins/X-Chain.xml $@
if [ "x$1" != "x--xml" ]
then
chmod a+x $SLICER_HOME/chains/chain.sh
$SLICER_HOME/chains/chain.sh > $SLICER_HOME/chains/result.log
fi
Chain Descriptor
- This is an XML file which contains all the information necessary to deterministically proxy into CLMs and:
- Build the new XML descriptor corresponding to the composition of the modules (removing chain arguments, applying namespaces, autogenerating certain values, etc.)
- Invoke them when necessary in sequential order respecting the argument piping instructions and the corresponding arguments for each modules' (using its namespaces).
- It must conform to the following schema:
<xs:schema xmlns:xs="http://www.w3.org/2001/XMLSchema" xmlns:clm="http://www.na-mic.org/clm" targetNamespace="http://www.na-mic.org/clm" elementFormDefault="qualified"> <xs:element name="modules"> <xs:complexType> <xs:sequence> <xs:element name="module" type="clm:module" minOccurs="2" maxOccurs="unbounded"/> </xs:sequence> </xs:complexType> </xs:element> <xs:complexType name="module"> <xs:sequence> <xs:element name="argument" type="clm:argument" minOccurs="0" maxOccurs="unbounded"/> </xs:sequence> <xs:attribute name="id" type="xs:string"/> </xs:complexType> <xs:complexType name="argument"> <xs:attribute name="name" type="xs:string"/> <xs:attribute name="value-ref" type="xs:string"/> </xs:complexType> </xs:schema>
- "Chain Descriptor" template example:
<?xml version="1.0" encoding="UTF-8"?> <modules xmlns="http://www.na-mic.org/clm" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.na-mic.org/schemas/chainCLM.xsd"> <module id="Module1"/> <module id="Module2"> <argument name="param2A" value-ref="Module1.param1A" /> <argument name="param2B" value-ref="Module1.param1B" /> </module> <module id="Module3"> <argument name="param3C" value-ref="Module1.param1C" /> <argument name="param3D" value-ref="Module2.param2D" /> </module> </modules>
Real Examples
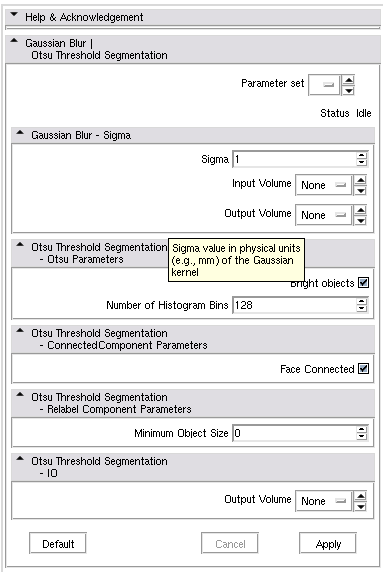
- Chaining modules GaussianBlurImageFilter and OtsuThresholdSegmentation:
<?xml version="1.0" encoding="UTF-8"?> <modules xmlns="http://www.na-mic.org/clm" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.na-mic.org/schemas/chainCLM.xsd"> <module id="GaussianBlurImageFilter"/> <module id="OtsuThresholdSegmentation"> <argument name="inputVolume" value-ref="GaussianBlurImageFilter.outputVolume" /> </module> </modules>
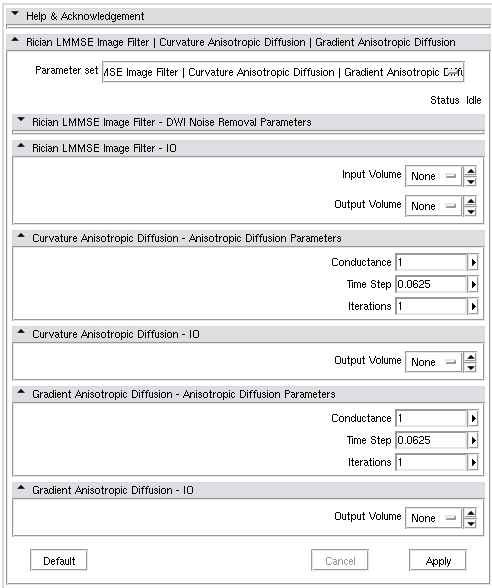
- Chaining modules dwiNoiseFilter, CurvatureAnisotropicDiffusion and GradientAnisotropicDiffusion:
<?xml version="1.0" encoding="UTF-8"?> <modules xmlns="http://www.na-mic.org/clm" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.na-mic.org/schemas/chainCLM.xsd"> <module id="dwiNoiseFilter"/> <module id="CurvatureAnisotropicDiffusion"> <argument name="inputVolume" value-ref="dwiNoiseFilter.outputVolume" /> </module> <module id="GradientAnisotropicDiffusion"> <argument name="inputVolume" value-ref="CurvatureAnisotropicDiffusion.outputVolume" /> </module> </modules>
- Chaining modules brainsegCLP, AffineRegistration, BSplineDeformableRegistration, ResampleVolume2, CortThickCLP
<?xml version="1.0" encoding="UTF-8"?> <modules xmlns="http://www.na-mic.org/clm" xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://www.na-mic.org/schemas/chainCLM.xsd"> <module id="brainsegCLP"/> <module id="AffineRegistration"/> <module id="BSplineDeformableRegistration"> <argument name="inputVolume" value-ref="AffineRegistration.ResampledImageFileName" /> <argument name="InitialTransform" value-ref="AffineRegistration.OutputTransform" /> <argument name="FixedImageFileName" value-ref="AffineRegistration.FixedImageFileName" /> <argument name="MovingImageFileName" value-ref="AffineRegistration.MovingImageFileName" /> </module> <module id="ResampleVolume2"> <argument name="InitialTransform" value-ref="BSplineRegistration.OutputTransform" /> </module> <module id="CortThickCLP"> <argument name="SegmentationInput" value-ref="brainseg.SegmentationFile" /> <argument name="ParcellationOption" value-ref="ResampleVolume2.outputVolume" /> </module> </modules>
Key Investigators
- UCSD: Marco Ruiz
- Isomics: Steve Pieper
- GE: Jim Miller