Difference between revisions of "On the performance of Slicer 2 and Slicer3 EM segmentation modules"
(Created page with '==Abstract== This page presents an experiment comparing the performance of Slicer 2.6 and Slicer 3.5 EM segmentation modules on no-human data. Sept, 09. === Experiment descripti…') |
|||
| Line 4: | Line 4: | ||
=== Experiment description === | === Experiment description === | ||
# DATA: | # DATA: | ||
| − | + | ||
| + | Experiment done on subject Oscar. This subject was used as the target in generating template, so the template and target are perfectly aligned. As a result, by choosing this subject, the effect of template-to-target mis-alignment in segmentation is removed. The resulted difference between Slicer 2 and 3 EM will purely reflect the performance of the segmentation modules. | ||
#Procedure | #Procedure | ||
| − | + | Data is first segmented in Slicer 3. The parameters are manually chosen (by selecting samples, etc.) for the best segmentation performance. The exact parameter are copied to perform the same segmentation in Slicer 2. | |
* EMsegmentation parameters in Slicer 3 : (Global prior | atlas weight) {log mean | log covariance} [label] | * EMsegmentation parameters in Slicer 3 : (Global prior | atlas weight) {log mean | log covariance} [label] | ||
| − | + | (Same EMsegmentation parameters in Slicer 2 : (prob | prob data weight) {mean | covariance} [label] ) | |
whole head | whole head | ||
Revision as of 20:50, 9 September 2009
Home < On the performance of Slicer 2 and Slicer3 EM segmentation modulesAbstract
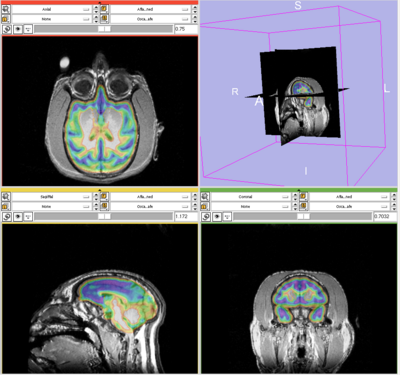
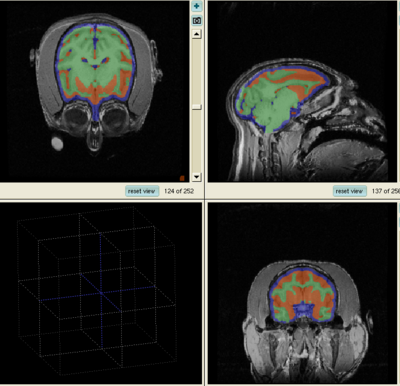
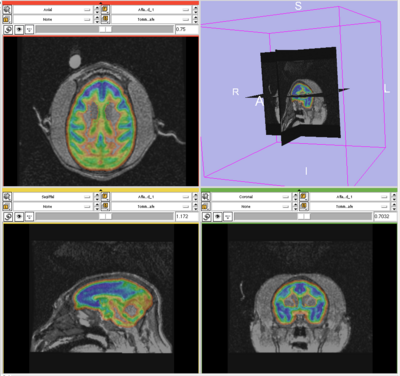
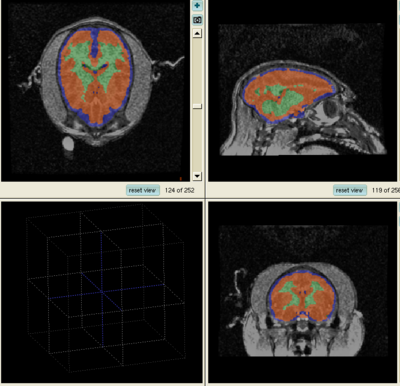
This page presents an experiment comparing the performance of Slicer 2.6 and Slicer 3.5 EM segmentation modules on no-human data. Sept, 09.
Experiment description
- DATA:
Experiment done on subject Oscar. This subject was used as the target in generating template, so the template and target are perfectly aligned. As a result, by choosing this subject, the effect of template-to-target mis-alignment in segmentation is removed. The resulted difference between Slicer 2 and 3 EM will purely reflect the performance of the segmentation modules.
- Procedure
Data is first segmented in Slicer 3. The parameters are manually chosen (by selecting samples, etc.) for the best segmentation performance. The exact parameter are copied to perform the same segmentation in Slicer 2.
- EMsegmentation parameters in Slicer 3 : (Global prior | atlas weight) {log mean | log covariance} [label]
(Same EMsegmentation parameters in Slicer 2 : (prob | prob data weight) {mean | covariance} [label] )
whole head
|_ECC (0.4 1) {2.9317, 1.7046} [l:0]
|_ICC (0.6 1)
|_GM (0.4 0.7)
| |_GMwoHPC (0.69 0.6) {3.5549,0.02454} [l:7]
| |_HP (0.07 0.6) {3.747,0.00214} [l:4]
| |_Caudate (0.14 0.6) {3.6354,0.00456} [l:5]
| |_Putamen (0.1 0.6) {3.7522,0.00456} [l:6]
|_WM (0.3 0.7) {3.9028,0.00548} [l:2]
|_CSF (0.3 0.7) {2.805,0.23349} [l:3]
Segmentation Results
Worst Segmentation: Tommy